Simulating Organogenesis in COMSOL Multiphysics®: Comparison of Methods for Simulating Branching Morphogenesis
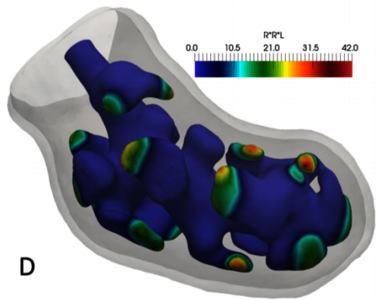
Organogenesis, the genesis of organs during embryonic development, is an active research field involving biologists, mathematicians and computational researchers. Fundamental questions of the formation of shape and the control of growth are still not answered. We focus on the development of the embryonic mouse lung and kidney. From all proposed mechanisms, a Turing Pattern-based framework as proposed in [1] predicts the experimentally determined growth field and thus locations of the emerging branches the best. Turing Patterns are spatio-temporal stable patterns emerging from a system of diffusing morphogens, e.g. proteins, which interact in a non-linear manner.
So far, we used COMSOL Multiphysics® and the Arbitrary Lagrangian-Eulerian (ALE) framework for solving the system of PDEs [2-4]. The complex deformation during lung growth limits the number of branch generations that can be simulated. We have previously proposed a Phase-Field-based approach to circumvent mesh deformations by tracking the surface of the emerging organ as the interface between two phases, still coupling the reaction-diffusion mechanism to the surface [5].
We now report a rigorous comparison to the ALE-based model. Moreover, we use biological data and infer parameter values for which the outgrowth of new branches reproduces the experimental observations. We use COMSOL Multiphysics® for all our simulations. The reaction-diffusion process of the Turing mechanism is modelled with the Coefficient Form PDE interface. We use the Level-Set interface to track the epithelial and mesenchymal border. This allows us to advect the interfaces separately simulating predicted outgrowth or incorporating measured biological data. Adaptive mesh refinement is employed to achieve a fine grained resolution in the region of the interfaces, while lowering the computational effort in the remaining domain.
References [1] D. Menshykau et al., "An interplay of geometry and signaling enables robust lung branching morphogenesis.", Development 141(23): 4526-4536 (2014)
[2] D. Menshykau and D. Iber, “Simulation Organogenesis in COMSOL: Deforming and Interacting Domains,” Proceedings of the 2012 COMSOL Conference in Milan (2012)
[3] Z. Karimaddini et al., “Simulating Organogenesis in COMSOL: Image-based Modeling.”, Proceedings of the 2014 COMSOL Conference in Cambridge (2014)
[4] P. Germann et al., “Simulating Organogenesis in COMSOL,” Proceedings of the 2011 COMSOL Conference in Stuttgart (2011)
[5] L. D. Wittwer et al., “Simulating Organogenesis in COMSOL: Phase-Field Based Simulations of Embryonic Lung Branching Morphogenesis”, Proceedings of the 2016 COMSOL Conference in Munich (2016)
Download
- wittwer_presentation.pdf - 30.11MB
- wittwer_paper.pdf - 1.93MB
- wittwer_abstract.pdf - 0.02MB
